Research interests
Bioinformatics, Artificial Intelligence and Data Analysis
- Development of new unsupervised and supervised machine learning methods
- Use of machine learning methods, including clustering and deep learning, for the analysis of biological and biomedical data
- Phylogenetic analysis
- Genomics
- Evolution and origin of SARS-CoV-2
- Analysis of CRISPR/CAS9 data
- Analysis of High Throughput Screening (HTS) data
Professor
|
Vladimir Makarenkov Full professor / Professeur titulaire Director of DESS in Bioinformatics at UQAM Département d'Informatique, Université du Québec à Montréal (UQAM) Regular member of LaCIM and affiliate member of Mila CV and publications |
Ph.D. / Doctorat
 |
Jael Champagne Gareau Master's (finished) and Ph.D student co-supervised with Éric Beaudry / étudiant à la maitrise (terminée) et au doctorat en informatique co-supervisé avec Éric Beaudry |
 |
Jocelyn Bédard Master's (finished) and Ph.D student / étudiant à la maitrise (terminée) et au doctorat en informatique 
|
 |
Mohamed Mediouni Master's (finished) and Ph.D student co-supervised with A.B. Diallo / étudiant à la maitrise (terminée) et au doctorat en informatique co-supervisé avec A.B. Diallo |
|
Zeinab Sherkatghanad Master's (finished) and Ph.D student / étudiante à la maitrise (terminée) et au doctorat en informatique |
Master's students / Maitrise
 |
Mohamed Achraf Bouaoune Master's student / étudiant à la maitrise en informatique 
|
 |
Yasmine Khelil Master's student / étudiante à la maitrise en informatique |
 |
Soumia Melek Master's student / étudiante à la maitrise en informatique |
Alumni
Postdocs
 |
Jeremy Charlier Postdoc Emploi actuel: AI Senior Manager at the National Bank of Canada / Montreal, Canada 
|
 |
Etienne Lord Postdoc Emploi actuel: Research Scientist in digital agronomy and A.I. at Agriculture and Agri-Food Canada / Government of Canada, St-Jean-sur-Richelieu, QC, Canada 
|
 |
Laszlo Szathmary Postdoc Emploi actuel: Professor at University of Debrecen, Hungary 
|
 |
Dmytro Kevorkov Postdoc Emploi actuel: Research Associate, Concordia University, Montreal 
|
 |
Alena Tsikhanovich Postdoc Emploi actuel: Senior Researcher, Famic Technologies Inc., Montreal 
|
 |
Alix Boc Postdoc Emploi actuel: Senior software developer at Code3, Montreal 
|
 |
Chérif Mballo Postdoc Emploi actuel: Conseiller Sénior en Modélisation Statistique, Gestion du Risque de Crédit chez Desjardins, Levis, QC |
 |
Jingxin Xie Postdoc Emploi actuel: Senior Research Engineer at Nuance Communications, Montreal 
|
 |
Andrei Gagarin Postdoc Emploi actuel: Research Associate, Royal Holloway, University of London 
|
Ph.D students / Étudiants au doctorat
 |
Moloud Abdar Ph.D student / étudiant au doctorat en informatique (UQAM and Deakin University, Australia) Emploi actuel: Postdoc at Deakin University, Australia |
 |
Nadia Tahiri Master's and Ph.D student / étudiante à la maitrise et au doctorat en informatique Emploi actuel: Professor at University of Sherbrooke, Canada / Professeur à l'Université de Sherbrooke, Canada 
|
 |
Matthieu Willems Master's and Ph.D student / étudiant à la maitrise et au doctorat en informatique Emploi actuel: Professor at Cegep André-Laurendeau, Montreal, Canada / Professeur au Cégep André-Laurendeau, Montréal, Canada |
|
Alexandre Gondeau Ph.D student / étudiant au doctorat en informatique Emploi actuel: Research engineer in Bioinformatics at Bordeaux Bioinformatics Center (CBiB), France 
|
 |
Alix Boc Master's and Ph.D student / étudiant à la maitrise et au doctorat en informatiqueWeb Site Emploi actuel: Senior software developer at Code3, Montreal 
|
 |
Dunarel Badescu Master's and Ph.D student / étudiant à la maitrise et au doctorat en informatique Emploi actuel: Bioinformatics specialist, McGill University 
|
 |
Plamen Dragiev Ph.D student (co-supervised with Robert Nadon, McGill University) / étudiant au doctorat en informatique Emploi actuel: Senior software developer, Ericsson, Montreal 
|
 |
Mehdi Layeghifard Ph.D student / étudiant au doctorat en sciences biologiques (co-supervised with Pedro Peres-Neto, Concordia University) Emploi actuel: Postdoctoral researcher, University of Toronto 
|
 |
Alpha Boubacar Diallo Master's and Ph.D student / étudiant à la maitrise et au doctorat en informatique Emploi actuel: Bioinformatics specialist, DNAnexus, Mountain View, California, USA 
|
 |
Dung Nguyen Ph.D student / étudiant au doctorat en informatique cognitive Emploi actuel: Senior software developer, Banque Nationale, Montreal 
|
Master's students / Étudiants à la maitrise*
 |
Nail Amine Chabane Master's student / étudiant à la maitrise en informatique Emploi actuel: Senior Data Scientist, National Bank of Canada 
|
 |
Reda Tighilt Master's student / étudiant à la maitrise en informatique Emploi actuel: Senior software developer, Synergie Médicale 
|
 |
Henry Xing Master's student co-supervised with S. Kembel / étudiant à la maitrise en informatique co-supervisé avec S. Kembel Emploi actuel: IT Specialist, S3 Technologies Inc. 
|
 |
Valérie Hay Master's student / étudiante à la maitrise en informatique Emploi actuel: Bioinformatician, Institut Cardiologie de Montréal 
|
 |
Nancy Badran Master's student / étudiante à la maitrise en informatique Emploi actuel: Data Analyst at Statistics Canada, Canada 
|
 |
Pablo Zentilli Master's student / étudiant à la maitrise en informatique Emploi actuel: Software developer, Analystik inc, Montréal 
|
 |
Tania Joly Master's student / étudiante à la maitrise en informatique Emploi actuel: Applications Analyste, Société de transport de Montréal 
|
 |
Adel Younes Master's student / étudiant à la maitrise en informatique Emploi actuel: Software developer, ST2i, Montréal 
|
|
Rabah Djema Master's student / étudiant à la maitrise en informatique |
|
Abdellah Mazouzi Master's student / étudiant à la maitrise en informatique |
Bachelor's students / Étudiants au Baccalauréat
 |
Bogdan Mazoure Bachelor student, McGill University - NSERC internship Emploi actuel: Research Scientist at Apple MLR 
|
 |
Iliya Reutsky Bachelor student, Concordia University Emploi actuel: Master's student at Concordia University and Software Developer at Genetec 
|
Latest publications
For the complete list of publications, please see my Google Scholar profile
Developed software
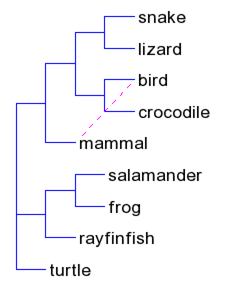 | T-REX web server (link) | The T-REX (tree and reticulogram reconstruction) web server allows the users to perform several popular methods of phylogenetic analysis as well as some new phylogenetic algorithms for inferring, drawing and statistical validation of phylogenetic trees and networks |
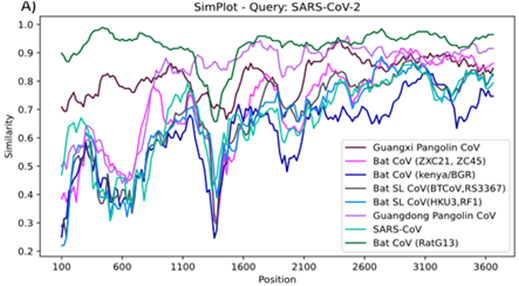 | SimPlot++ (link) | Multi-platform application for representing sequence similarity and detecting recombination |
 | DuneScan (link) | Deep uncertainty estimation for skin cancer (DuneScan web server) |
 | CRISPR-Cas9 (data and program repository) (link) | CRISPR-Cas9 benchmark data and program repository for on- and off-target experiments |
 | Armadillo workflow platform (link) | Armadillo is a workflow platform dedicated to phylogenetic as well as general bioinformatics analysis. A number of important computational biology tools have been included in the first release. The Armadillo platform is open-source and allows users to develop their own modules and/or integrate existing computer applications |
 | HTS Corrector (link) | HTS Corrector software is designed to examine and visualize HTS data and correct experimental HTS assays |
 | Optimal Variable Weighting (link) | Optimal variable weighting for ultrametric and additive clustering and k-means partitioning (with Pierre Legendre, Université de Montréal) |
 | Linear and Polynomial RDA and CCA (link) | Linear and polynomial canonical analysis (with Pierre Legendre, Université de Montréal) |
 | Robinson and Foulds distance (link) | Program for computation of the Robinson and Foulds topological distance between two phylogenetic trees (with Pierre Legendre, Université de Montréal) |




