Logiciels développés
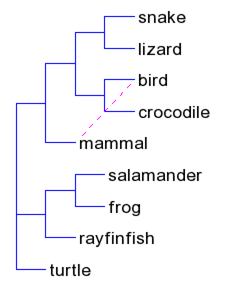 | T-REX web server (link) | The T-REX web server allows users to perform several popular methods of phylogenetic analysis as well as some new phylogenetic applications for inferring, drawing and validating phylogenetic trees and networks which we have developed.
Standalone software Program T-REX (tree and reticulogram reconstruction) |
 | T-REX web server (link) | The T-REX (tree and reticulogram reconstruction) web server allows the users to perform several popular methods of phylogenetic analysis as well as some new phylogenetic algorithms for inferring, drawing and statistical validation of phylogenetic trees and networks |
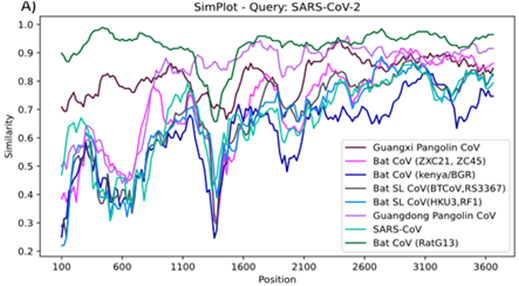 | SimPlot++ (link) | Multi-platform application for representing sequence similarity and detecting recombination |
 | DuneScan (link) | Deep uncertainty estimation for skin cancer (DuneScan web server) |
 | CRISPR-Cas9 (data and program repository) (link) | CRISPR-Cas9 benchmark data and program repository for on- and off-target experiments |
 | Armadillo workflow platform (link) | Armadillo is a workflow platform dedicated to phylogenetic as well as general bioinformatics analysis. A number of important computational biology tools have been included in the first release. The Armadillo platform is open-source and allows users to develop their own modules and/or integrate existing computer applications |
 | HTS Corrector (link) | HTS Corrector software is designed to examine and visualize HTS data and correct experimental HTS assays |
 | Optimal Variable Weighting (link) | Optimal variable weighting for ultrametric and additive clustering and k-means partitioning (with Pierre Legendre, Université de Montréal) |
 | Linear and Polynomial RDA and CCA (link) | Linear and polynomial canonical analysis (with Pierre Legendre, Université de Montréal) |
 | Robinson and Foulds distance (link) | Program for computation of the Robinson and Foulds topological distance between two phylogenetic trees (with Pierre Legendre, Université de Montréal) |